Exercise 2: Monte-Carlo
The Law of Large Numbers and Monte-Carlo Estimation.
Write a function which generates a sample of \(10\) observations \(iid\) from a Gaussian distribution (with mean \(m=2\) and standard deviation \(s=3\), i.e. a variance of \(9\)) and that returns the variance estimate over this sample (using the
var()function).var_est <- function(n = 10) { s <- rnorm(n, mean = 2, sd = 3) return(var(s)) }Use the the Monte-Carlo method to estimate the variance of the distribution generating this sample, by using multiple realizations (e.g. 5) of the standard-deviation estimate implemented above. Do it again with 1,000 realizations, thus illustrating the law of large number convergence.
# Monte-Carlo method nMC <- 5 # Monte Carlo sample size varMC <- numeric(nMC) for (i in 1:nMC) { varMC[i] <- var_est(n = 10) } mean(varMC) # LLN estimate## [1] 6.653662nMC <- 1000 # Monte Carlo sample size for (i in 1:nMC) { varMC[i] <- var_est(n = 10) } mean(varMC) # LLN estimate## [1] 9.126242When the sample size increases, the estimator becomes more precise. This illustrates the Law of Large Numbers.
Let’s now program a Monte-Carlo estimate of \(\pi\approx 3,1416\)
Program a function
roulette_coordwhich has only one argumentngrid(representing the number of different outcomes possible on the roulette used) whose default is35, generating the two coordinates of a point (between \(0\) and \(35\)) as a vector. Use theRfunctionsample(whhose help page is accessible through the command?sample). The function will return the vector of the 2 coordinatesxandygenerated this way.roulette_coord <- function(ngrid = 35) { x <- sample(x = 0:ngrid, size = 1) y <- sample(x = 0:ngrid, size = 1) return(c(x, y)) }Thanks to the formula to compute the distance bewteen 2 points: \(d = \sqrt{(x_1 - x_2)^2 + (y_1 - y_2)^2}\), program a function computing the distance to the origin (here has coordinates \((\frac{ngrid}{2}, \frac{ngrid}{2})\)) that checks if the computed distance is less than the unit disk radius (\(R = \frac{ngrid}{2}\)). This function, called for instance
inside_disk_fun(), will have 2 arguments: the vectorpcontaining the coordinates of the points on the one hand, and the integerngridon the other hand. It will return a boolean value (TRUEorFALSE) indicating the point is inside the disk.inside_disk_fun <- function(p, ngrid = 35) { d <- sqrt((p[1] - ngrid/2)^2 + (p[2] - ngrid/2)^2) return(d <= ngrid/2) }The surface ratio between the disk (radius \(\frac{ngrid}{2}\)) and the square (side length \(ngrid\)) is equal to \(\frac{\pi}{4}\). This means that the probability of sampling a point inside the disk rather than outside is \(\frac{\pi}{4}\). Using this result, a Monte Carlo estimate of \(\pi\) can be implemented by computing the average number of time sampled points fall inside the disk multiplied by 4. Program such a function with their only input being a boolean vector of size \(n\) (the number of sampled points), which is
TRUEif the point is indeed inside the disk andFALSEotherwise.piMC <- function(in_disk) { return(4 * mean(in_disk)) }Using the code below, generate 200 points and plot the data generated. What is the corresponding Monte Carlo estimate of \(\pi\) ? Change
npointsand comment. How could the estimation be improved (ProTip: tryngrid <- 1000andnpoints <- 5000) ?# Grid size (resolution) ngrid <- 35 # Monte Carlo sample size npoints <- 200 # Points generation pp <- matrix(NA, ncol = 2, nrow = npoints) for (i in 1:nrow(pp)) { pp[i, ] <- roulette_coord(ngrid) } # Estimate pi in_disk <- apply(X = pp, MARGIN = 1, FUN = inside_disk_fun, ngrid = ngrid) piMC(in_disk) # Plot first we initialise an empty plot with the right size using argument plot(x = pp[, 1], y = pp[, 2], xlim = c(0, ngrid), ylim = c(0, ngrid), axes = 0, xlab = "x", ylab = "y", type = "n") ## we tick the x and then y axes from 1 to ngrid axis(1, at = c(0:ngrid)) axis(2, at = c(0:ngrid)) ## we add a square around the plot box() ## we plot the grid (using dotted lines thanks to the argument `lty = 3`) onto ## which the points are sample for (i in 0:ngrid) { abline(h = i, lty = 3) abline(v = i, lty = 3) } ## we add the sampled points lines(x = pp[, 1], y = pp[, 2], xlim = c(0, ngrid), ylim = c(0, ngrid), xlab = "x", ylab = "y", type = "p", pch = 16) ## we add the circle display x.circle <- seq(0, ngrid, by = 0.1) y.circle <- sqrt((ngrid/2)^2 - (x.circle - ngrid/2)^2) lines(x.circle, y = y.circle + ngrid/2, col = "red") lines(x.circle, y = -y.circle + ngrid/2, col = "red") ## finally we color in red the points sampled inside the disk lines(x = pp[in_disk, 1], y = pp[in_disk, 2], xlim = c(0, ngrid), ylim = c(0, ngrid), xlab = "x", ylab = "y", type = "p", pch = 16, col = "red", cex = 0.7)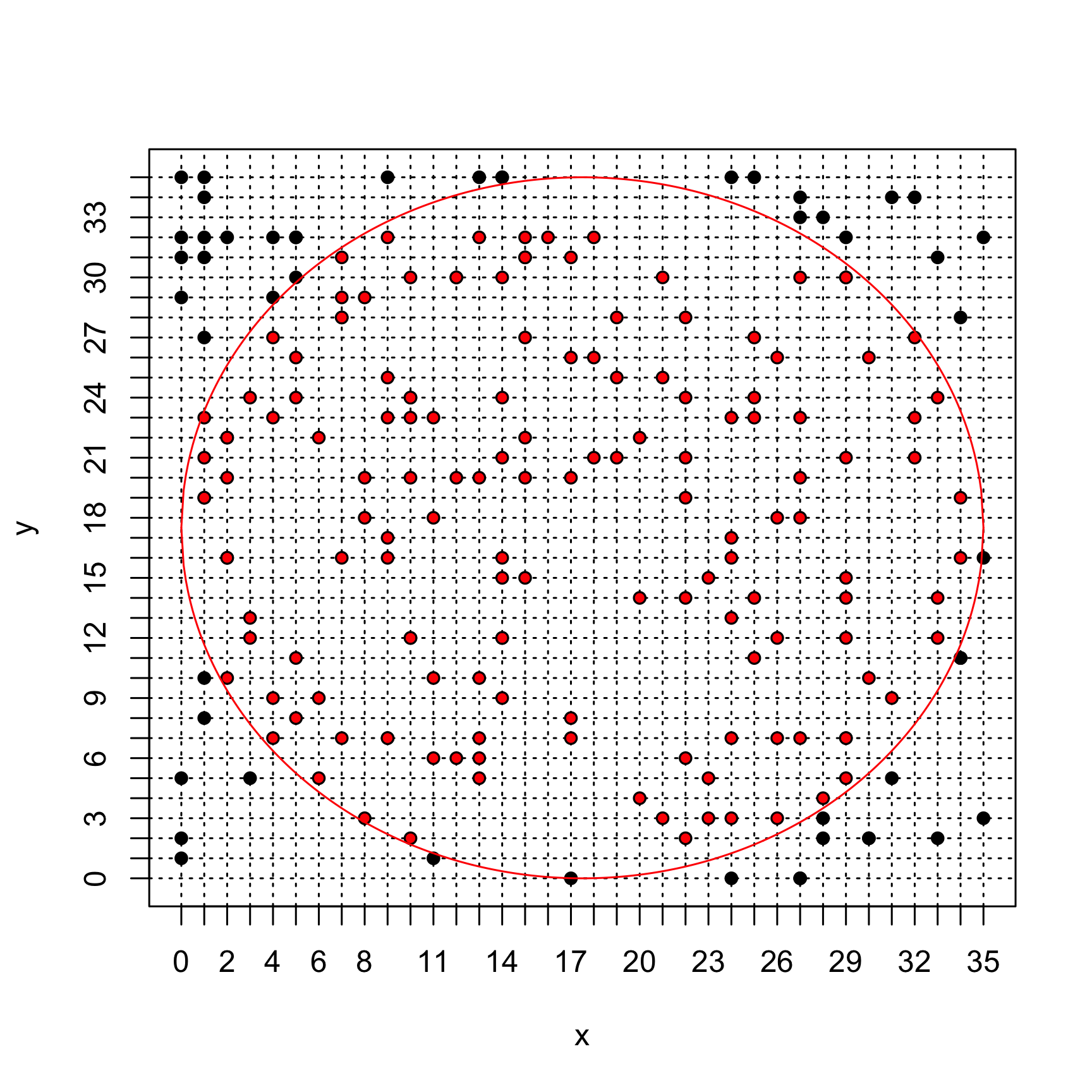
When the sample size increase, the Monte Carlo estimator becomes more precise (LLN). However, if the grid is too coarse, \(\widehat{\pi}\) is underestimated (underestimating the disk surface by missing the bits near the edge). Therefore, increasing the number of points on the grid also improves the precision of the Monte Carlo estimation.