Exercise 6: Post-mortem analysis of an under-powered randomized trial
The randomized clinical trial EOLIA1 evaluated a new treatment for severe acute respiratory distress syndrome (severe ARDS) by comparing the mortality rate after 60 days among 249 patients randomized between a control group (receiving conventional treatment, i.e. mechanical ventilation) and a treatment group receiving extracorporeal membrane oxygenation (ECMO) — the new treatment studied. A frequentist analysis of the data concluded to a Relative Risk of death of
Goligher et al. (2019) 2 performed a Bayesian re-analysis of these data, further exploring the evidence and how it can be quantified and summarized with a Bayesian approach.
| Control | ECMO | |
|---|---|---|
| 125 | 124 | |
| number of deceased at 60 days | 57 | 44 |
Write the Bayesian model used by Goligher et al. (2019).
I) Question of interest:
Is the Relative Risk of death under ECMO compared to the conventional mechanical treatment less than one ?
II) Sampling model:
Let
III) Priors:
NB:
NB: One can also define a sampling model at the individual level:
LetWrite the corresponding BUGS model, and save it into a
.txtfile (for instance calledgoligherBUGSmodel.txt)As we have seen above, there are two equivalent ways of defining the sampling model:- either at the population level with a Binomial likelihood,
- or at the individual level with a Bernoulli likelihood
# Population model model{ # Sampling model zcontrol~dbin(pc, ncontrol) zecmo~dbin(RR*pc, necmo) # Prior logRR~dunif(-17, 17) # SD is approximately 10 pc~dunif(0,1) #probability of death in the control group # Re-parameterizations RR <- exp(logRR) ARR <- pc - RR*pc }# Individual model model{ # Sampling model for (i in 1:ncontrol){ ycontrol[i]~dbern(pc) } for (i in 1:necmo){ yecmo[i]~dbern(RR*pc) } # Prior logRR~dunif(-17, 17) # SD is approximately 10 pc~dunif(0,1) #probability of death in the control group # Re-parameterizations RR <- exp(logRR) ARR <- pc - RR*pc }First create two binary data vectors
ycontrolandyecmo(orycontrolandyecmothat are either1or0, using therep()R function if you prefer the individual model), to encode the observations from the data table above. Then uses thejags.model()andcoda.samples()to replicate the estimation from Goligher et al. (2019) (ProTip: use the functionwindow()to remove the burn-in observation from the output of thecoda.samplesfunction.)#Individual data ycontrol <- c(rep(0, 125-57), rep(1, 57)) yecmo <- c(rep(0, 124-44), rep(1, 44)) #sampling library(rjags) goligher_jags_indiv <- jags.model(file = "goligherBUGSmodel_indiv.txt", data = list("ycontrol" = ycontrol, "ncontrol" = length(ycontrol), "yecmo" = yecmo, "necmo" = length(yecmo) ), n.chains = 3)## Compiling model graph ## Resolving undeclared variables ## Allocating nodes ## Graph information: ## Observed stochastic nodes: 249 ## Unobserved stochastic nodes: 2 ## Total graph size: 260 ## ## Initializing modelres_goligher_indiv <- coda.samples(model = goligher_jags_indiv, variable.names = c('pc', 'RR', 'ARR'), n.iter = 40000) #postprocessing res_goligher_burnt_indiv <- window(res_goligher_indiv, start=21001) # remove burn-in for Markov chain convergence #NB: by default coda.samples() performs 1000 first burnin iterations that are automatically removed from the output but till indexed. #Population data zcontrol <- 57 zecmo <- 44 #sampling goligher_jags_pop <- jags.model(file = "goligherBUGSmodel_pop.txt", data = list("zcontrol" = zcontrol, "ncontrol" = 125, "zecmo" = zecmo, "necmo" = 124 ), n.chains = 3)## Compiling model graph ## Resolving undeclared variables ## Allocating nodes ## Graph information: ## Observed stochastic nodes: 2 ## Unobserved stochastic nodes: 2 ## Total graph size: 13 ## ## Initializing modelres_goligher_pop <- coda.samples(model = goligher_jags_pop, variable.names = c('pc', 'RR', 'ARR'), n.iter = 40000) #post-processing res_goligher_burnt_pop <- window(res_goligher_pop, start=21001) # remove burn-in for Markov chain convergence #NB: by default coda.samples() performs 1000 first burnin iterations that are automatically removed from the output but till indexed.Check the convergence, and then comment the estimate results (ProTip: look at the effective sample size with the
effectiveSize()Rfunction).effectiveSize(res_goligher_burnt_pop)## ARR RR pc ## 14879.42 16214.78 15998.81plot(res_goligher_burnt_pop)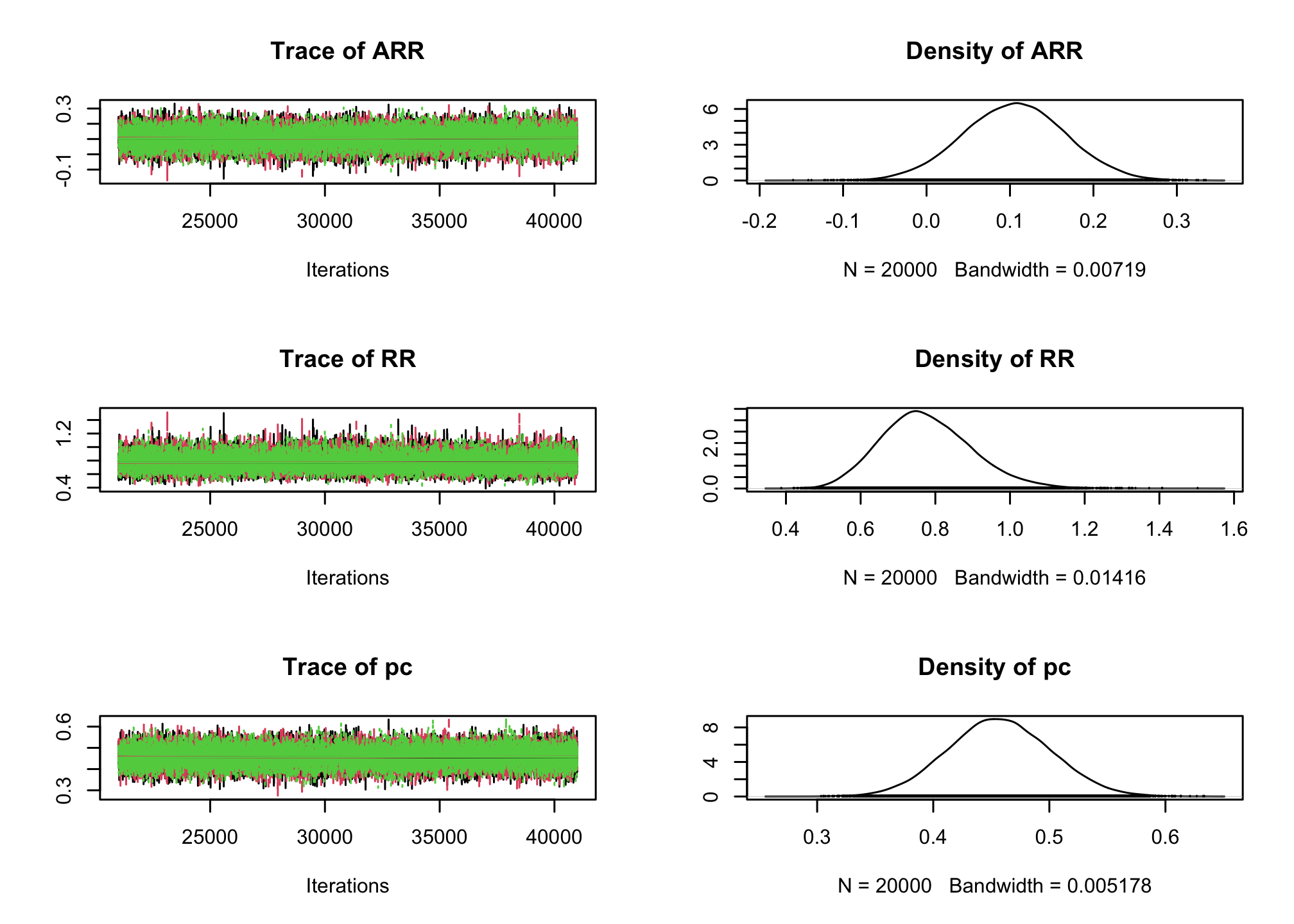
gelman.plot(res_goligher_burnt_pop)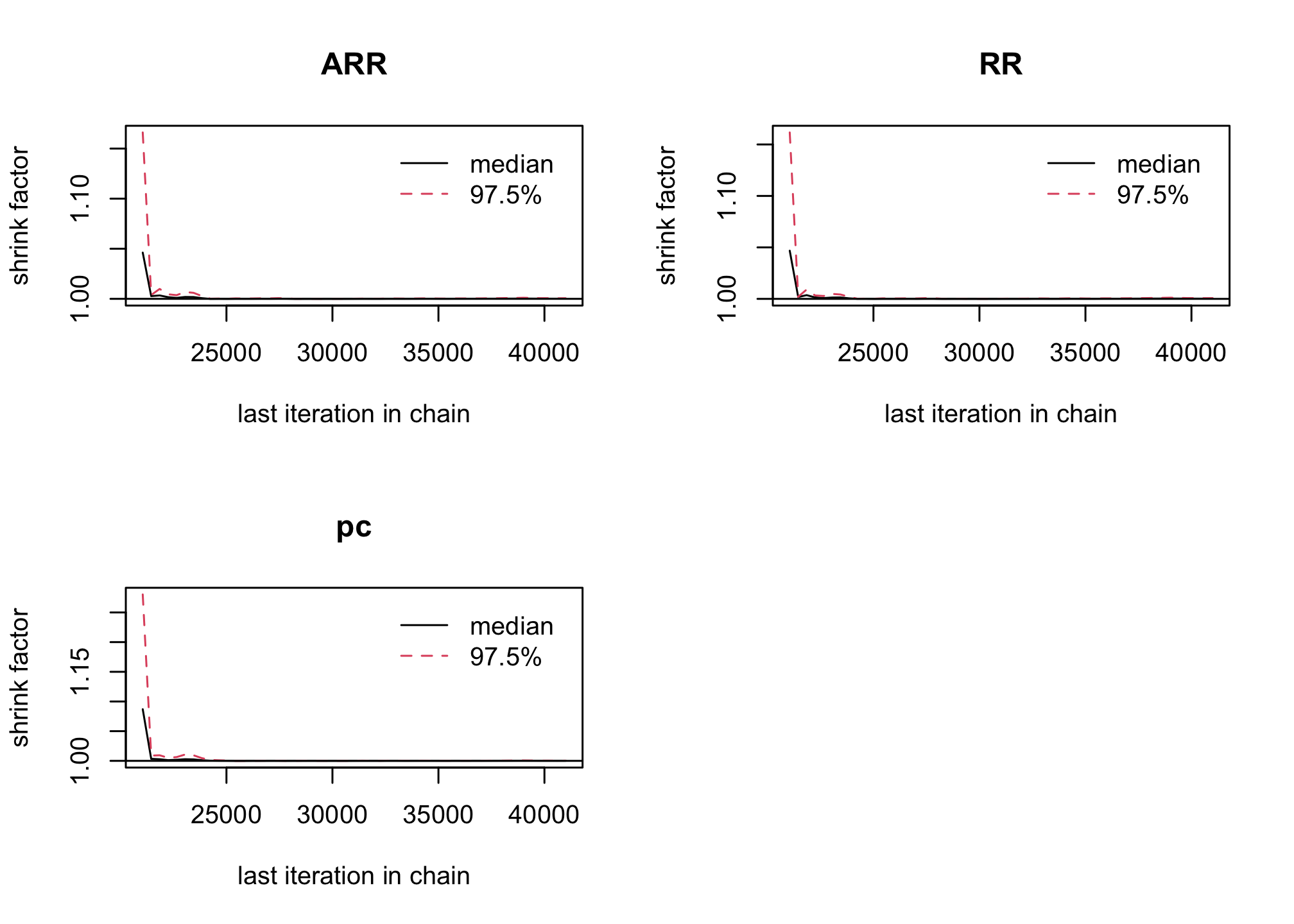
acfplot(res_goligher_burnt_pop)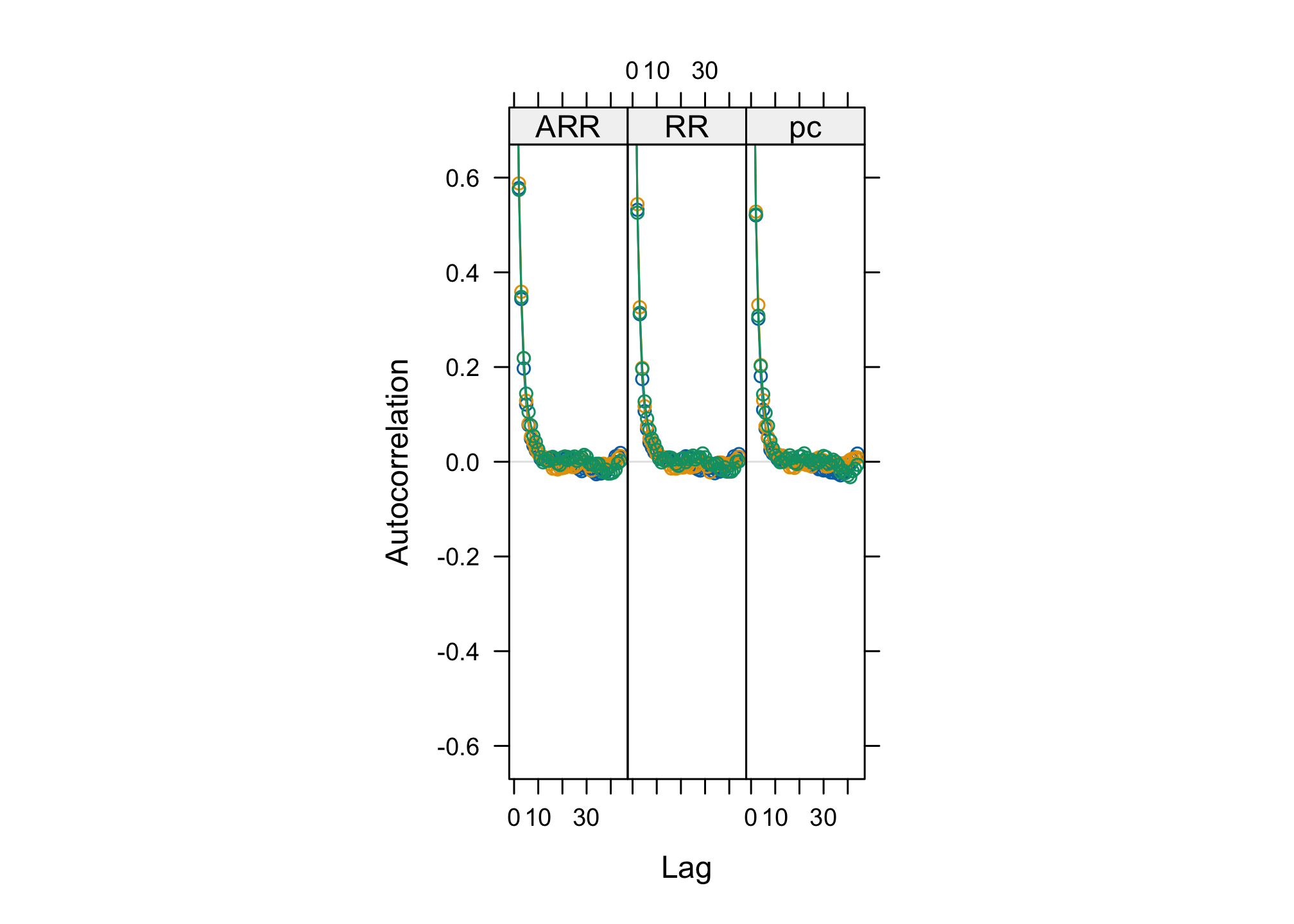
par(mfrow=c(3, 2)) cumuplot(res_goligher_burnt_pop, ask=FALSE, auto.layout = FALSE)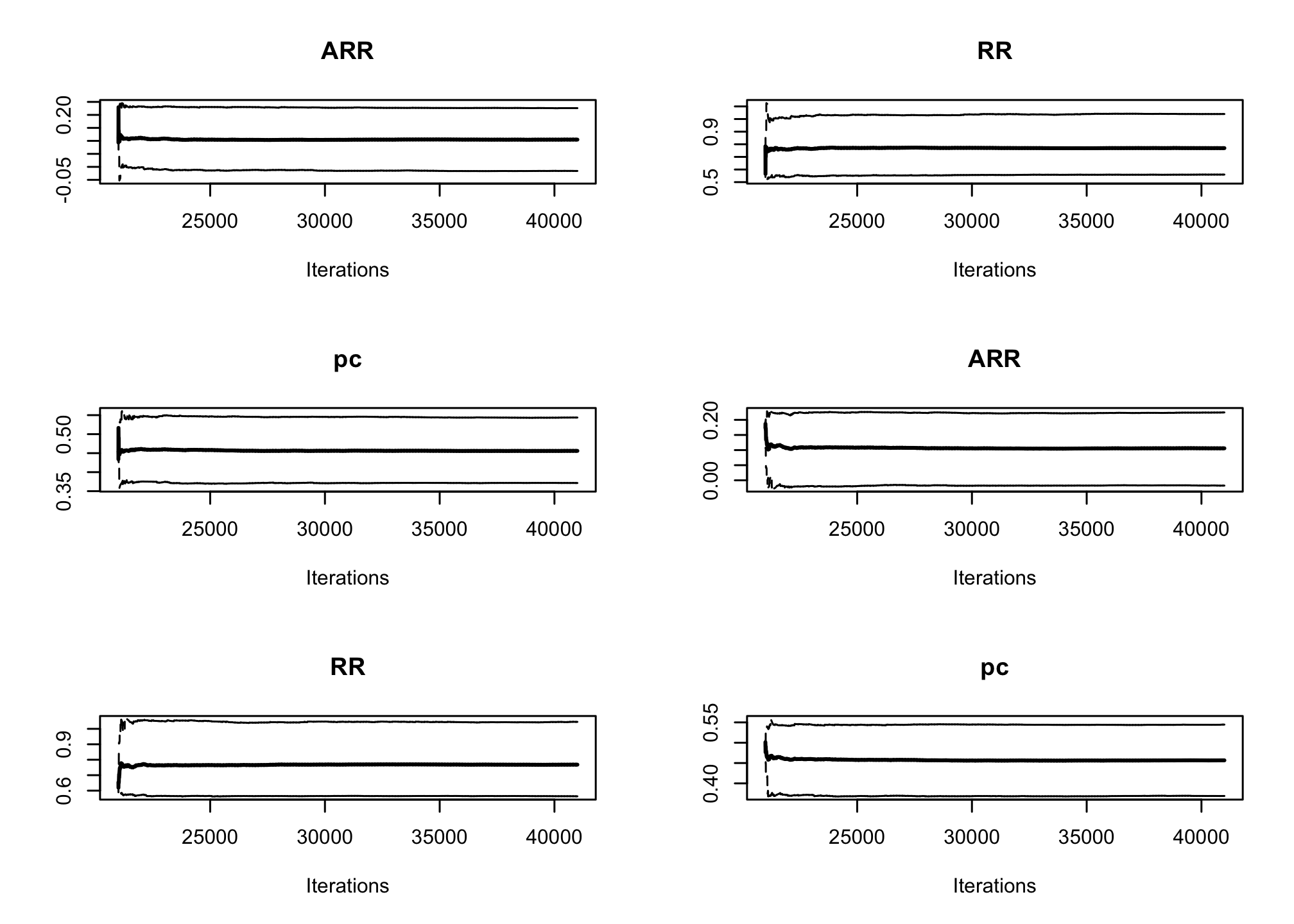
par(mfrow=c(1, 1))
summary(res_goligher_burnt_pop)## ## Iterations = 21001:41000 ## Thinning interval = 1 ## Number of chains = 3 ## Sample size per chain = 20000 ## ## 1. Empirical mean and standard deviation for each variable, ## plus standard error of the mean: ## ## Mean SD Naive SE Time-series SE ## ARR 0.1054 0.06125 0.0002500 0.0005024 ## RR 0.7767 0.12163 0.0004965 0.0009555 ## pc 0.4570 0.04411 0.0001801 0.0003491 ## ## 2. Quantiles for each variable: ## ## 2.5% 25% 50% 75% 97.5% ## ARR -0.01526 0.06381 0.1059 0.1467 0.2251 ## RR 0.56161 0.69183 0.7681 0.8534 1.0392 ## pc 0.37146 0.42709 0.4565 0.4867 0.5442summary(res_goligher_burnt_indiv)## ## Iterations = 21001:41000 ## Thinning interval = 1 ## Number of chains = 3 ## Sample size per chain = 20000 ## ## 1. Empirical mean and standard deviation for each variable, ## plus standard error of the mean: ## ## Mean SD Naive SE Time-series SE ## ARR 0.1047 0.06090 0.0002486 0.0004963 ## RR 0.7779 0.12151 0.0004961 0.0009552 ## pc 0.4566 0.04378 0.0001787 0.0003409 ## ## 2. Quantiles for each variable: ## ## 2.5% 25% 50% 75% 97.5% ## ARR -0.01617 0.06406 0.1052 0.1461 0.2223 ## RR 0.56563 0.69258 0.7697 0.8534 1.0411 ## pc 0.37144 0.42684 0.4564 0.4861 0.5422# shortest 95% Credibility interval: HDInterval::hdi(res_goligher_burnt_pop)## ARR RR pc ## lower -0.01362207 0.5488287 0.3706276 ## upper 0.22654858 1.0198553 0.5432404 ## attr(,"credMass") ## [1] 0.95# posterior porbability of RR <1: (answering our question !) head(res_goligher_burnt_pop[[1]]) # first chain## Markov Chain Monte Carlo (MCMC) output: ## Start = 21001 ## End = 21007 ## Thinning interval = 1 ## ARR RR pc ## [1,] 0.09361565 0.7843224 0.4340537 ## [2,] 0.14264251 0.7237648 0.5163806 ## [3,] 0.22962327 0.5650301 0.5279061 ## [4,] 0.24341419 0.5206346 0.5077842 ## [5,] 0.24490921 0.5392786 0.5315777 ## [6,] 0.19667001 0.6005747 0.4923825 ## [7,] 0.05165148 0.8946283 0.4901838head(res_goligher_burnt_pop[[2]]) # second chain## Markov Chain Monte Carlo (MCMC) output: ## Start = 21001 ## End = 21007 ## Thinning interval = 1 ## ARR RR pc ## [1,] 0.1733616 0.6541119 0.5012072 ## [2,] 0.1867682 0.6191842 0.4904425 ## [3,] 0.1963678 0.5879483 0.4765609 ## [4,] 0.1842940 0.6361408 0.5064982 ## [5,] 0.1225179 0.7411831 0.4733769 ## [6,] 0.1233054 0.7291038 0.4551757 ## [7,] 0.1029334 0.7727277 0.4529078mean(c(sapply(res_goligher_burnt_pop, "[", , "RR"))<1)## [1] 0.9564833Change to a more informative prior using a Gaussian distribution for the log(RR), centered on log(0.78) and with a standard deviation of 0.15 in the log(RR) scale (i.e. a precision of
1/(0.15^2)## [1] 44.44444logRR~dnorm(log(0.78), 45)
Alain Combes et al. “Extracorporeal Membrane Oxygenation for Severe Acute Respiratory Distress Syndrome,” New England Journal of Medicine 378, no. 21 (2018): 1965–1975, doi:10.1056/NEJMoa1800385.↩︎
Ewan C. Goligher et al. “Extracorporeal Membrane Oxygenation for Severe Acute Respiratory Distress Syndrome and Posterior Probability of Mortality Benefit in a Post Hoc Bayesian Analysis of a Randomized Clinical Trial,” JAMA 320, no. 21 (2018): 2251, doi:10.1001/jama.2018.14276.↩︎